Coronaviruses (CoVs) – SARS-CoV & SARS-CoV-2 [2019-nCoV]
Coronaviruses (CoVs) are enveloped non-segmented positive-sense RNA viruses infecting humans and vertebrates. They are classified into four types (genus), α-CoV, β-CoV, γ-CoV and δ-CoV. They can infect the respiratory, gastrointestinal, hepatic and central nervous systems of humans and many wild animals. The family of Coronaviridae constantly circulates within the human population and mainly causes mild respiratory diseases. Recently, a new severe acute respiratory syndrome β-coronavirus called SARS-CoV-2 (or 2019-nCoV) has emerged, which causes an epidemic of acute respiratory syndrome called coronavirus human disease 2019 or COVID-19. Typical clinical symptoms of these patients are dry cough, fever, breathing difficulties, headache and pneumonia. Disease onset may result in progressive respiratory failure, heart tissue damage and even death.
SARS-CoV-2 shares 79.5% sequence identity with SARS-CoV and is 96.2% identical at the genome level to the bat coronavirus BatCoV RaTG133, suggesting it had originated in bats. The coronaviral genome encodes four major structural proteins: the Spike (S) protein, Nucleocapsid (N) protein, Membrane/Matrix (M) protein and the Envelope (E) protein (see Figure 1). The SARS Envelope (E) protein plays a role in viral budding and virion envelope morphogenesis. The SARS Membrane/Matrix (M) protein is one of the major structural viral proteins. It is an integral membrane protein involved in the budding of the viral particles and interacts with SARS Spike (S) protein and the Nucleocapsid (N) protein. The N protein contains two domains, both of which bind the virus RNA genome via different mechanisms.
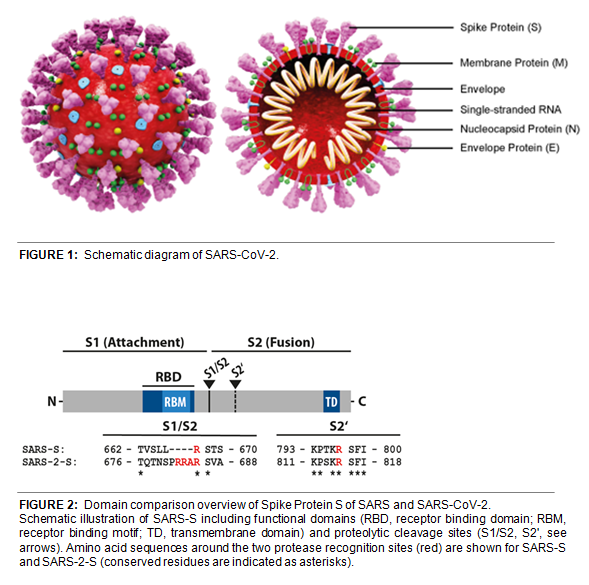
The CoV Spike (S) protein assembles as a trimer and plays the most important role in viral attachment, fusion and entry. It is composed of a short intracellular tail, a transmembrane anchor, and a large ectodomain that consists of a receptor-binding S1 subunit and a membrane-fusing S2 subunit (see Figure 2, adapted from Cell, Hofmann, et al. (2020)). The S1 subunit contains a receptor binding domain (RBD), which binds to the cell surface receptor angiotensin-converting enzyme 2 (ACE2) present at the surface of epithelial cells.
LITERATURE REFERENCES:
- The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak – an update on the status: Y.R. Guo, et al.; Mil. Med. Res. 7, 11 (2020) (Review)
- SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor: M. Hoffmann, et al.; Cell 181, 731 (2020)
- Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine: W. Tai, et al.; Cell Mol. Immunol. 17, 613 (2020)
- Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2: R. Yan, et al.; Science 367, 1444 (2020)
- Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein: A.C. Walls, et al.; Cell 181, 281 (2020) (Review)
Originally posted on adipogen.com/covid-19
Caltag Medsystems is the distributor of Adipogen products in the UK and Ireland. If you have any questions about these products, please contact us.
Related Blogs:
- ACE2 & Functional Receptor for SARS Coronaviruses
- Biological Therapeutic Strategies against SARS-CoV-2 and COVID-19
- COVID-19 Diagnostics
- Antiviral Compounds – Potential Small Molecule Therapeutics Against COVID-19
- Cytokine Storm and Severity of COVID-19
