SARS-CoV-2 (COVID-19) Spike S2 Antibody [P1A6]
| Code | Size | Price |
|---|
| PSI-SD9785-0.02mg | 0.02mg | £166.00 |
Quantity:
| PSI-SD9785-0.1mg | 0.1mg | £531.00 |
Quantity:
Prices exclude any Taxes / VAT
Overview
Host Type: Llama
Antibody Isotype: sdAb
Antibody Clonality: Recombinant Antibody
Antibody Clone: P1A6
Regulatory Status: RUO
Target Species: SARS-CoV-2
Application: Enzyme-Linked Immunosorbent Assay (ELISA)
Shipping:
Dry Ice
Storage:
SARS-CoV-2 (COVID-19) Spike S2 Antibody should be stored in working aliquots at -20˚C, stable for up to one year. As with all antibodies care should be taken to avoid repeated freeze thaw cycles. Antibodies should not be exposed to prolonged high temperatures.
Images
Documents
Further Information
Additional Names:
SARS-CoV-2 (COVID-19) Spike S2 Antibody: Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), Surface Glycoprotein, Spike protein
Background:
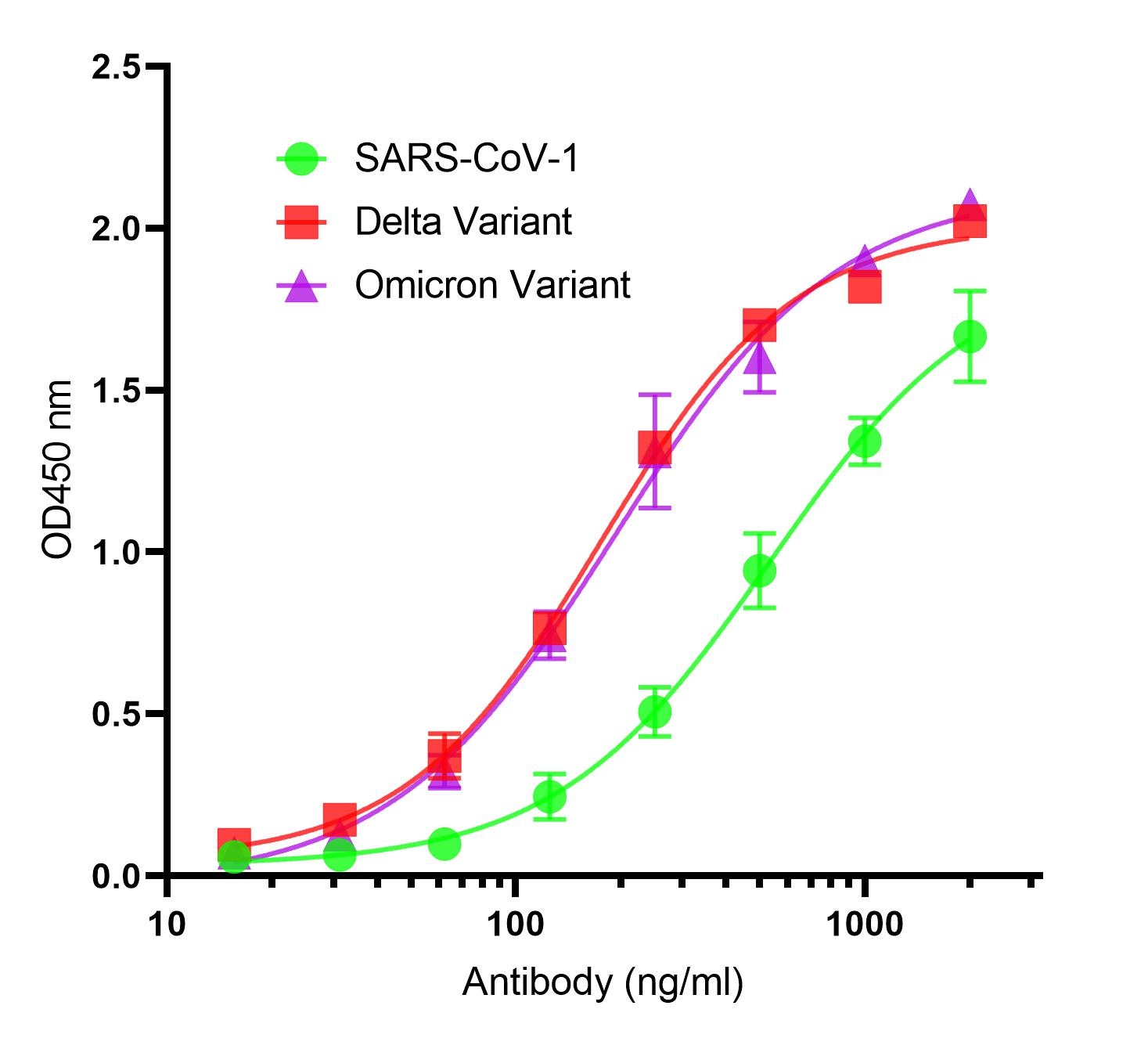
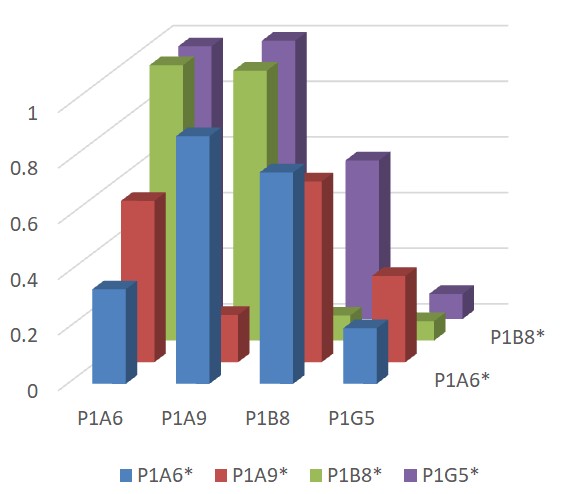
As SARS-CoV-2 Spike S2 protein is more conserved among coronaviruses than Spike S1 protein, antibodies against spike S2 protein may react with more Coronaviruses, serving as a pan-coronvirus antibody. The As SARS-CoV-2 Spike S2 protein is more conserved among coronaviruses than Spike S1 protein, antibodies against spike S2 protein may react with more Coronaviruses, serving as a pan-coronvirus antibody. The functional characteristics analysis revealed that P1A6 has excellent binding activity to spike trimer of SARS-CoV-1, and SARS-CoV-2 variants (delta and omicron). Biotinylated P1A6 can still bind spike trimer of SARS-CoV-2. In addition, P1A6 and another three spike S2 sdAbs (P1G5, P1A9, and P1B8) belong to different families based on their sequence alignment, and epitope mapping experiment indicated that they may bind to different epitopes with different affinity to the different strains in the binding ELISA. Our data showed that these spike S2 sdAbs bind spike trimer of both SARS-CoV-2 and SARS-CoV-1, suggesting that these spike S2 sdAbs have potentials as a novel and efficient tool suitable for pan-coronavirus studies as well as for identification of various coronaviruses and their variants from clinical samples. functional characteristics analysis revealed that P1A6 has excellent binding activity to spike trimer of SARS-CoV-1, and SARS-CoV-2 variants (delta and omicron). Biotinylated P1A6 can still bind spike trimer of SARS-CoV-2. In addition, P1A6 and another three spike S2 sdAbs (P1G5, P1A9, and P1B8) belong to different families based on their sequence alignment, and epitope mapping experiment indicated that they may bind to different epitopes with different affinity to the different strains in the binding ELISA. Our data showed that these spike S2 sdAbs bind spike trimer of both SARS-CoV-2 and SARS-CoV-1, suggesting that these spike S2 sdAbs have potentials as a novel and efficient tool suitable for pan-coronavirus studies as well as for identification of various coronaviruses including their variants from clinical samples.
Buffer:
SARS-CoV-2 (COVID-19) Spike S2 Antibody is supplied in PBS.
Concentration:
batch dependent
Conjugate:
Unconjugated
Immunogen:
SARS-CoV-2 Spike S2 protein containing C-terminal His Tag. The protein was expressed in human 293 cells (HEK293). It contains amino acids Ser 686 - Pro 1213.
NCBI Gene ID #:
43740568
NCBI Official Name:
S
NCBI Official Symbol:
S
NCBI Organism:
surface glycoprotein
Physical State:
Liquid
Protein Accession #:
QHD43416
Protein GI Number:
1791269090
Purification:
SARS-CoV-2 (COVID-19) Spike S2 Antibody is affinity chromatography purified via Nickel column. Antibody is supplied as a His-tagged purified protein.??It also contains a myc-tag for detection.
Research Area:
Infectious Disease,COVID-19
User NOte:
Optimal dilutions for each application to be determined by the researcher.